ORIGINAL
Genetic variability of broodstocks of restocking programs in Brazil
Variabilidad genética de lotes de reproductores de programas de repoblación en Brasil
Nelson Lopera-Barrero,1* Ph.D, Ed Lima,1 Zoot, Luiz Filho,2 Ph.D, Elenice Goes,2 M.Sc, Pedro Castro,2 Zoot, Aline Zardin,2 M.Sc, Angela Poveda-Parra,1 Ph.D, Ricardo Ribeiro2, Ph.D.
1Universidade Estadual de Londrina, Departamento de Zootecnia, Programa de Pós-Graduação em Ciência Animal, Rod. Celso Garcia Cid Pr 445 hm 380 Campus Universitário, Londrina, Brasil.
2Universidade Estadual de Maringá, Departamento de Zootecnia, Programa de Pós-Graduação em Zootecnia, Avenida Colombo 5790 Jardim Universitário, Maringá, Brasil.
*Correspondence: nmlopera@uel.br
Received: August 2014; Accepted: February 2015.
ABSTRACT
Objective. The aim of this study was evaluate the genetic diversity of the following broodstocks: piapara (Leporinus elongatus), dourado (Salminus brasiliensis), jundiá (Rhamdia quelen) and cachara (Pseudoplatystoma fasciatum) already useful for restocking programs in the Paranapanema, Iguaçu and Paraná Brazilian Rivers. Materials and methods. Samples from the caudal fin of 122 fish were analyzed. DNA was extracted by NaCl protocol. PCR products were separated by a horizontal agarose gel electrophoresis. The fragments were visualized by staining with ethidium bromide. Results. The amplification of 25 primers generated different fragments in studied species that allowed characterizing 440 fragments of 100-2900 bp. High percentage of polymorphic fragments (66.67 to 86.29), Shannon index (0.365 to 0.486) and genetic diversity of Nei (0.248 to 0.331) were detected. Conclusions. The level of genetic variability in the broodstocks was adequate for allowing their use in restocking programs in the studied Rivers. However, periodical monitoring studies of genetic variability in these stocks, the mating system, reproductive system and general management must be made to guarantee the preservation of wild populations.
Key words: Genetic conservation, Leporinus elongatus, Pseudoplatystoma fasciatum, Rhamdia quelen, Salminus brasiliensis (Source: DeSC, CAB Thesaurus).
RESUMEN
Objetivo. El objetivo de este estudio fue evaluar la diversidad genética de los siguientes lotes de reproductores: piapara (Leporinus elongatus), dourado (Salminus brasiliensis), jundiá (Rhamdia quelen) y cachara (Pseudoplatystoma fasciatum) utilizados para programas de repoblación en los ríos brasileños Paranapanema, Iguaçu y Paraná. Materiales y métodos. Muestras de aleta caudal de 122 peces fueron analizadas. El ADN fue extraído por el protocolo de NaCl. Los productos de PCR fueron separados por electroforesis horizontal en gel de agarosa. Los fragmentos fueron visualizados por marcación con bromuro de etidio. Resultados. La amplificación de los 25 iniciadores produjo diferentes fragmentos en las especies estudiadas que permitieron caracterizar 440 fragmentos de 100 a 2900 pb. Fueron detectados un alto porcentaje de fragmentos polimórficos (66.67 a 86.29), de índice de Shannon (0.365 a 0.486) y de diversidad genética de Nei (0.248 a 0.331). Conclusiones. El nivel de variabilidad genética en los lotes de reproductores fue adecuado para su utilización en programas de repoblación en los ríos estudiados. Sin embargo, estudios de monitoreo periódico de la variabilidad genética en esos lotes, del sistema de cruzamiento, del sistema reproductivo y del manejo general deben ser realizados para garantizar la preservación de las populaciones naturales.
Palabras clave: Conservación genética, Leporinus elongatus, Pseudoplatystoma fasciatum, Rhamdia quelen, Salminus brasiliensis (Fuente: DeSC, CAB Thesaurus).
INTRODUCTION
In the last seven years, fishing of the world aquatic organisms has pointed out toward decreases and stabilization. In 2005 and 2010, only 92.478.416 and 88.970.124 tons were, respectively, captured, which means a reduction of 3.508.292 tons. In 2011, in contrast, 93.494.340 tons had 4.524.216 tons higher than in 2010 (1). The fish farming, otherwise, has been more significant. For example, in 2005 and 2010 the growth was higher than the catching, when there was an increase from 57.815.836 to 78.028.539 tons, respectively, representing a net output of 20.212.703 tons. In 2011, furthermore, 83.675.661 tons of fish were raised; an output of 5.647.122 tons higher than in 2010 (1).
In Brazil, based on data from the Aquaculture and Fisheries Ministry (2), the fishing exploitation had an increase from 785.366 in 2010 to 803.270 tons in 2011, or 2.3%. Otherwise, the fish farming increased from 479.398 in 2010 to 628.704 tons in 2011, or 31.1%. These data suggest an overlap of fish farming on fishing exploitation in the next years as well as detected in other segments of the animal production (3). The piapara (Leporinus elongatus), dourado (Salminus brasiliensis), jundiá (Rhamdia quelen) and cachara (Pseudoplatystoma fasciatum) species has been highlighted because they are present in several Brazilian basins (4) with several factors encouraging their farming as the good growth and performance, the easy reproduction and the rusticity (5-8). These factors have impacted the Brazilian market of fish. For example, in 2011 were produced 4309.3 and 1747.3 tons of Rhamdia and Leporinus (2).
Other factors, however, as the water pollution, deforestation, habitat destruction, overfishing, introduction of exotic species and river damming (9,10) has been reducing the wild populations of these freshwater fish in Brazil. Among the several decisions made to mitigate the reduction of the wild populations of fish, we highlight the restocking programs (11). Despite they have been applied in Brazil for decades, researches about their genetic and environmental efficiency have still to be done (12). Based on Kalinowski et al (13), the goals of such programs for endangered species have been to avoid the complete losses of gene clusters, preserve as much as possible the genetic diversity and reach the first step without compromise the survivorship of the populations in a long term. Many programs, however, were unsuccessful because the mismanagement of parental stocks (11) and the genetic non-monitoring. Thus, the genetic evaluation of the stocks for restocking has to be done. The molecular markers have been one of the most useful methods to approach the genetic diversity and the population structure of the various species and fish lines (14,15).
The aim of the current study was to evaluate the genetic diversity of the piapara (Leporinus elongatus), dourado (Salminus brasiliensis), jundiá (Rhamdia quelen) and cachara (Pseudoplatystoma fasciatum) broodstocks in restocking programs applied in the Paranapanema, Iguaçu and Paraná Brazilian Rivers.
MATERIAL AND METHODS
Study site and sample collection. Caudal fins samples from broodstocks of the species Leporinus elongatus (30 samples), Salminus brasiliensis (30 samples), Rhamdia quelen (36 samples) and Pseudoplatystoma fasciatum (26 samples) were collected. These broodstocks are originally from collections carried in the Paranapanema River - São Paulo State (S 22° 26’ and 47° 54’ W - L. elongatus and S. brasiliensis), Iguacu River - Paraná State (S 25° 40’ and 54° 26’ W - R. quelen) and Paraná river - Parana State (S 23° 16’ and 53° 43’ W - P. fasciatum), Brazil. All these fish will be used to compose the restocking programs in these respective rivers.
Experimental procedure. The DNA was extracted using the protocol described by Lopera-Barrero et al (16). The DNA extracted was quantified in the Shimadzu spectrophotometer using the absorbance of 260 nm. The samples were diluted to 10 ng/µL, and the DNA quality was evaluated in 1% agarose gel electrophorese using a buffer TBE 1X (500 mM Tris-HCl, 60 mM boric acid and 83 mM EDTA) at 70 V for one hour. The gel was visualized under UV radiation after treatment with ethidium bromite (0.5 mg/ml) for one hour. Thereafter, the image was photographed using the program Kodak EDAS (Kodak 1D Image Analysis 3.5, EUA).
The DNA was amplified in volume of reaction of 15 µL using the buffer 1X Tris KCl, 2 mM of MgCl2, 0.46 µM of primer, 0.2 mM of every dNTP, one unity of Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, EUA), and 10 ng of target DNA.
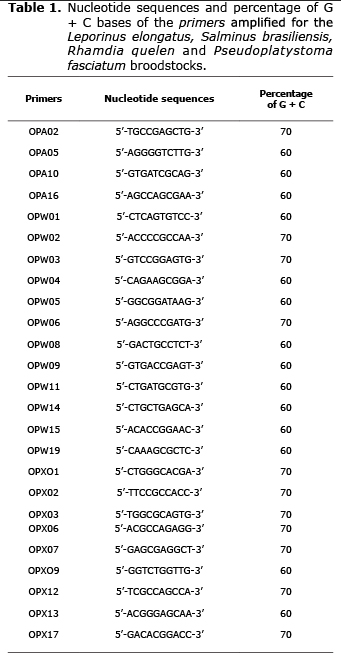
The reactions were amplified in Eppendorf Mastercycler Gradient (EUA), and the cycles were based on the species. L. elongatus and S. brasiliensis had the DNA denaturized at 96°C for 5 min and soon after was conducted 40 cycles of 1 min at 94°C, 1 min and 30 s annealing the primer at 36°C and extension at 72°C for 2 min. Thereafter, the final extension was at 72°C for 7 min. Otherwise, R. quelen had DNA denaturized at 92°C for 4 min and soon after was conducted 40 cycles of 1 min at a 92°C, 1 min of annealing at 40°C and 2 min at 72°C for extension. Soon after, was did the final extension at 72°C for 5 min. Finally, P. fasciatum had amplification programmed for 40 cycles, with one initial step of denaturation at 95°C for 5 min and the final step at 72°C for 7 min. Every cycle had 1 min at 94°C, 1 min at 36°C and two min at 72°C. Twenty five primers from the Kits OPA, OPW and OPX (Operon Technologies Ltd., Valencia, USA) with 10 bases for every species were tested, when those of best definition and reproducibility were chosen (Table 1).
The amplified products were separated in agarose gel at 1.5% using horizontal electrophoresis. Fifteen microliters of amplified product in conjunction with 2 µL of sample buffer (40% sucrose and 0.25% bromophenol blue) were inserted into the gel. This electrophoresis was carried out in buffer TBE 0.5X (45 mM Tris-Borate and 1 mM EDTA) at 70 V for 4 hours. The quantification and amplification gel were visualized under UV radiation after ethidium bromide dyeing (0.5 µg/mL) for 1 hour. The images were photographed using the program EDAS (Kodak 1D Image Analysis 3.5, New York, USA).
Computer software and statistical analysis. The size of the fragments was estimated by comparing them to the standard ladder of 100 bp (Invitrogen, Carlsbad, USA). The presence or absence of fragments with similar molecular size was used to build a similarity matrix based on the Jaccard coefficient where 1 was the presence and 0 was the absence. The percentage of polymorphic fragments (95% criterion), the Shannon’s index and the genetic diversity of Nei were calculated by the PopGene 1.31 program (17).
RESULTS
The 25 primers (Table 1) had different amplification standards for every fish species. A total of 440 fragments from 100 bp (OPW14-Pseudoplatystoma fasciatum) to 2900 bp (OPW03 - Leporinus elongatus) were amplified. The total number of fragments and number of polymorphic fragments were different among the species. In L. elongatus, 107 in 124 fragments were polymorphic; S. brasiliensis had 76 in 105; R. quelen had 128 in 134 fragments, being the highest number of fragments and percentage detected in the current experiment. Finally, P. fasciatum had 71 in 77 fragments (Table 2).
About the percentage of polymorphic fragments high values were found for all the species (66.67% to 86.29%). The Shannon index values were similarly high. The results from the genetic diversity of Nei showed high estimates, ranging from 0.248 (S. brasiliensis) to 0.331 (R. quelen) (Figure 1).
DISCUSSION
The total number of fragments found is consistent to the conclusions of Telles et al. (18) that determined the most important number of fragments obtained (at least 50) than the number of primers used for the calculation of genetic diversity.
The percentage of polymorphic fragments, Shannon index and genetic diversity of Nei parameters indicated high genetic variability that is essential for a good performance of restocking programs. Losses alleles of disease resistance and reduction in the adaptation capacity are consequences of dropping in the genetic variability (14). Literature data from molecular markers with the same studied species are displayed in the table 3. References to P. fasciatum were not found and only one manuscript to R. quelen was reported. Not all the cited references available had evaluated the same parameters investigated in the current experiment.
About the percentage of polymorphic fragments (PPF) and Shannon index (SI) from other manuscripts (Table 3), just the percentage of polymorphic fragments of R. quelen found by Lidani et al (19) was higher than was found in the current investigation. Both parameters (PPF and SI) were slightly lower in all manuscripts about L. elongatus and much more pronounced for S. brasiliensis. Most of the samples evaluated by these authors were collected in Paranapanema River. Working with S. brasiliensis, Lopes et al (20) stated that the genetic variability found in their stocks was the consequence of the small population in the sites where these fish were collected (Canoas complex, Paranapanema River). Thus, despite the river from where was collected the specimens of L. elongatus and S. brasiliensis had been the same the founder effect may have had influence on the genetic variability. Based on Kang et al (21), the small effective number of parental fish in the stock formation may eliminate low frequent alleles, and consequently reducing the genetic variability. This was the factor reported by Lopera Barrero et al (22) which induced high variability in Prochilodus lineatus because the stocks investigated were formed by fish populations from different Rivers. Similarly, Gomes et al (23) found low genetic variability in a group of seven S. brasiliensis with 33.33% of polymorphic fragments and Shannon index of 0.174, the lowest values found in these groups.
The genetic diversity of Nei is a parameter which determines the level of population differentiation. Based on the values obtained in this study, it can be affirmed that there was high heterogeneity between stocks, suggesting that they do not have a common origin. For this parameter, the values expressed in table 3 were classified from low to very high. Based on low values of genetic diversity (0.086), Ramos et al (10) stated that groups collected different periods in the transposition ladder of the UHE in Canoas I and Canoas II were part of only one population with several sub-populations. Similar conclusion was found by Lopez et al (20) to observe a value of 0.018. They also stated that this and other groups were structured in just one population from Capivara reservoir
The efficiency of restocking programs has also to be highlighted beyond proper genetic variability, i.e. the program intensity, number of parents using in the mating, mating system, general management, and the genetic differentiation between wild and broodstock populations.
Evaluating the intensity of restocking of Salvelinus fontinalis on the genetic diversity of wild populations, Marie et al (24) stated that in the lakes with higher intensity had higher levels of diversity, likely because of the introduction of domestic alleles. However, Lopera Barrero et al (22) reported the importance of genetic monitoring of populations to avoid losses in genetic variability (outbreeding depression), while preventing the introduction of genetic material com high genetic differentiation in relation to the wild population.
On the other hand, showing the negative effect of impropriated number of parents, Machado-Schiaffino et al (25) reported the variability losses in juveniles of Atlantic salmon (Salmo salar L.) from restocking programs in comparison to wild populations likely because the bottleneck effect caused by the reduced number used in breeding matings. In contrast, Lopera-Barrero et al (26) stabilized the variability using 24 parental fish (12 males and 12 females) of Brycon orbignyanus showing the positive effect of adequate number of parents, despite the semi-natural reproductive system that also had influence in maintain the genetic variability.
Another factor with influence in the genetic variability of fish stocks has been the mating system. Gomes et al (14) found higher estimates of genetic variability in the progeny of S. brasiliensis than in the parental fish using the semi-natural reproductive system. When evaluating the paternity of progeny Brycon orbignyanus in semi-natural and extrusion reproductive systems, Lopera Barrero et al (27) found multiple paternities and differentiated reproductive contribution in both systems. The authors, however, suggest more experiments to confirm the reproductive efficiency of semi-natural system in maintaining the genetic variability in other species.
The management of the offspring can also influence the genetic variability of restocking programs. Almeida et al (28) found that instead of releasing each offspring lot separately into the water, mixing specimens produced in the various fry stocks before releasing them in the river would be more feasible. The restocked population will have a genetic structure closer to wild populations. On the other hand, Rodriguez-Rodriguez et al (29) suggested that in the progeny aimed at restocking programs a first genetic analysis must be carried in the larval stage (3 days), which will provide a general overview of the new genetics generation. Then, at 60 or 90 days, depending on environmental conditions of the ecosystem should be performed a second analysis to objectively determine the final genetic variability with which individuals will be released into the river. This analysis should be performed to determine which individuals will be released and determine the true feasibility of restocking.
Finally, high genetic differentiation between wild and broodstocks can induce important gene losses associated with local adaptation. Similarly, González-Wangüemert et al (30) found high genetic differentiation between stocks of Diplodus sargus (for restocking the Castellamare gulf, Italy) and samples from the wild population. Based on these authors, this differentiation was likely verified by periodic inclusion of wild individuals from other places and the management adopted in captivity.
Based on the current results, the level of genetic variability in the L. elongatus, S. brasiliensis, R. quelen and P. fasciatum broodstocks are therefore adequate for allowing their use in restocking programs in the Paranapanema, Iguaçu and Paraná Rivers. However, periodical monitoring of the genetic variability of these stocks, the mating system, proper reproductive system and general management should be studied to guarantee the preservation of these wild populations.
Acknowledgements
The authors are thanks to the Duke Energy International for providing part of the biological material, and the researchers, technicians and students who supported the current experiment.
REFERENCES
1. Fao - Food and Agriculture Organization of the United Nations. FAO yearbook. Fishery and Aquaculture Statistics, 2010. Rome, 2013.
2. Mpa - Ministério da Pesca e Aquicultura. Boletim Estatístico da Pesca e Aquicultura, 2011. Brasília, 2013.
3. Fao - Food and Agriculture Organization of the United Nations. FAO yearbook. Fishery and Aquaculture Statistics, 2009. Rome, 2011.
4. Lopera-Barrero NM, Ribeiro RP, Povh JA, Vargas-Mendez LD, Poveda-Parra AR. Produção de Organismos Aquáticos: uma visão geral no Brasil e no mundo. Guaíba: Agrolivros; 2011.
5. Crepaldi DV, Faria PMC, Teixeira EA, Ribeiro LP, Costa AAP, Melo DC. O surubim na aquacultura do Brasil. Rev Bras Reprod Anim 2006; 30:150-158.
6. Graça WJ, Pavanelli CS. Peixes da planície de inundação do alto rio Paraná e áreas adjacentes. Maringá: EDUEM; 2007.
7. Kutter MT, Bemvenuti MA, Moresco A. Feeding strategy of the jundiá Rhamdia quelen (Siluriformes, Heptapteridae) in costal lagoons of southern Brazil. Acta Sci Biol Sci 2009; 31:41-47.
8. Gomes PC, Ribeiro RP, Lopera-Barrero NM, Povh JA, Vargas L, Sirol RN. Diversidade genética de três estoques de piapara (Leporinus elongatus), utilizando RAPD. Acta Sci Anim Sci 2008; 30:241-247.
9. Agostinho AA, Pelicice FM, Gomes LC. Dams and the fish fauna of the Neotropical region: impacts and management related to diversity and fisheries. Braz J Biol 2008; 68:1119-1132.
10. Ramos JVB, Sodré LMK, Orsi ML, Almeida FS. Genetic diversity of the species Leporinus elongatus (Teleostei: Characiformes) in the Canoas Complex - Paranapanema River. Neotrop Ichthyol 2012; 10:821-828.
11. Lopera Barrero NM. Conservation of Brycon orbignyanus natural populations and stocks for their reproductive, genetic, environmental sustainability: A model for species threatened with extinction. Cienc Inv Agr 2009; 36:191-208.
12. Povh JA, Ribeiro RP, Sirol RN, Streit JR DP, Moreira HLM, Siewerdt F et al. Microsatellite Analysis of the Parental Contribution of Piaractus mesopotamicus to the Production of Offspring in the Semi-natural System of Reproduction. Braz Arch Biol Technol 2010; 53:389-396.
13. Kalinowski ST, Doornik DMV, Kozfkay CC, Waples RS. Genetic diversity in the Snake River sockeye salmon captive broodstock program as estimated from broodstock records. Conserv Genet 2012; 13:1183-1193.
14. Gomes PC, Ribeiro RP, Sirol RN, Lopera Barrero NM, Moreira HLM, Povh JA et al. Diversidade genética de dourado utilizado em programas de repovoamento no rio Paranapanema. Pesq Agropec Bras 2011; 46:167-173.
15. Lopera Barrero NM, Povh JA, Fornari DC, Rodriguez Rodriguez M del P, Santos SCA, Ribeiro RP. Genetic diversity of Prochilodus lineatus stocks using in the stocking program of Tietê River, Brazil. Rev MVZ Córdoba 2013; 18:3759-3766.
16. Lopera-Barrero NM, Povh JA, Ribeiro RP, Gomes PC, Jacometo CB, Lopes TS. Comparison of DNA extraction protocols of fish fin and larvae samples: modified salt (NaCl) extraction. Cienc Inv Agr 2008a; 35:65-74.
17. Yeh FC, Boyle TYZ, Xiyan JM. POPGENE Version 131: Microsoft Window-based freeware for population genetic analysis. (Consulta en internet) Alberta (Accesado mayo de 2014). 1999. URL Disponible en: http://www.ualberta.ca/~fyeh/popgene.pdf.
18. Telles MPC, Monteiro MSR, Rodrigues FM, Soares TN, Resende LV, Amaral AG et al. Marcadores RAPD na análise de divergência genética entre raças de bovinos e número de locos necessários para a estabilidade da divergência estimada. Cien Anim Bras 2001; 2:87-95.
19. Lidani KCF, Lima JR, Torres RA, Gabriel JE, Madeira HMF, Carneiro PCF. Variabilidade genética de um estoque cativo de Jundiá (Rhamdia quelen). Rev Acad 2006; 4:47-53.
20. Lopes CM, Almeida FS, Orsi ML, Britto SGC, Sirol RN, Sodré LMK. Fish passage ladders from Canoas Complex – Paranapanema River: evaluation of genetic structure maintenance of Salminus brasiliensis (Teleostei: Characiformes). Neotrop Ichthyol 2007; 5:131-138.
21. Kang JH, Noh JK, Kim JH, Lee JH, Kim HC, Kim KK et al. Genetic relationship between broodstocks of olive flounder, Paralichthys olivaceus (Temminck and Schlegel) using microsatellite markers. Aqua Res 2006; 37:701-707.
22. Lopera Barrero NM, Ribeiro RP, Povh JA, Gomes PC, Vargas L, Oliveira SN. Caracterización genética de lotes de peces usados en programas de repoblamiento y su importancia en la conservación genética en la piscicultura. Zootecnia Trop 2008b; 26:515-522.
23. Gomes PC, Lopera Barrero NM, Vargas L, Streit Jr DP, Povh JA, Sirol RN et al. Genetic diversity of Salminus brasiliensis (Characiformes: Characidae) collected in the passage ladder of the Canoas I hydropower plant in the Paranapanema River, Brazil. Semin-Cienc Agrar 2013; 34:1421-1432.
24. Marie D, Bernatchez L, Garant D. Loss of genetic integrity correlates with stocking intensity in brook charr (Salvelinus fontinalis). Mol Ecol 2010; 19:2025-2037.
25. Machado-Schiaffino G, Dopico E, Garcia-Vazquez E. Genetic variation losses in Atlantic salmon stocks created for supportive breeding. Aquacult 2007; 264:59-65.
26. Lopera-Barrero NM, Vargas L, Sirol RN, Ribeiro RP, Povh JA, Mangolin CA. Caracterização genética de Brycon orbignyanus utilizando o sistema seminatural. Arq Bras Med Vet Zootec 2010; 62:184-191.
27. Lopera-Barrero NM, Reyez Alvarez CA, Rodriguez-Rodriguez MP, Povh JA, Vargas L, Streit Jr DP et al. Diversidade genética e paternidade de progênies de Brycon orbignyanus obtidas por diferentes sistemas reprodutivos. Semin-Cienc Agrar 2014; 35:541-554.
28. Almeida FS, Lopes CM, Orsi ML, Sirol RN, Sodré LMK. Genetic monitoring by RAPD markers for repopulation programs of Salminus brasiliensis (Pisces, Characiformes). Acta Sci Anim Sci 2013; 35:119-126.
29. Rodriguez-Rodriguez MP, Lopera Barrero NM, Ribeiro RP, Povh JA, Vargas L, Sirol RN et al. Diversidad genética de piracanjuba usada en programas de repoblación con marcadores microsatélites. Pesq Agropec Bras 2010; 45:53-63.
30. Gonzáles-Wangüemert M, Fernández TV, Pérez-Rufaza A, Giacalone M, D’anna G, Badalamenti F. Genetic considerations on the introduction of farmed fish in marine protected areas: The case of study of white seabream restocking in the Gulf of Castellammare (Southern Tyrrhenian Sea). J Sea Res 2012; 28:41-48.